Donà Erika
- Details
- Hits: 1962
 Research scientist
Research scientist
c/o Università di Milano - Bicocca
Via Raoul Follereau, 3
20854 Vedano al Lambro (MB)
Tel. 02 64488 397
This email address is being protected from spambots. You need JavaScript enabled to view it.
Scientific career path
I studied Medical Biotechnology and Molecular Medicine at the University of Milan (Italy), taking a master curriculum focussed on neuroscience. I then got interested in development and went to Darren Gilmour’s lab at EMBL (Heidelberg, Germany) for my PhD. There I studied how a group of cells in the zebrafish embryo is able to self-direct its long-distance movement by shaping its own chemokine gradient. During my postdoc in Greg Jefferis’ lab at the MRC-LMB (Cambridge, UK) I combined my interests in development and neuroscience by studying the developmental and molecular basis of sex-specific differences in the fly brain and their impact on animal behaviour.
Research summary
Building circuit specificity throughout development
The architecture of neural circuits is responsible for controlling information processing and animal behaviour. It results from the integrated action of cell specification and differentiation, neurite patterning and synapse formation. What are the developmental and molecular mechanisms that coordinate this complex multi step process?
To tackle this question we are employing a combination of genetics, high-throughput single-cell transcriptomics and connectomics in Drosophila melanogaster. This model system offers a number of experimental advantages: it has a tractable number of neurons in comparison to mammalian systems, it is characterised by high morphological stereotypy between individuals and a plethora of tools are available to obtain genetic access to individual neurons repeatedly across animals. Additionally, its complete connectome has recently become available, namely a comprehensive map of all the neurons and their synapses in the nervous system, which offers the unique possibility to link single-cell level gene expression profiles through development to end-point neuroanatomy, connectivity and function.
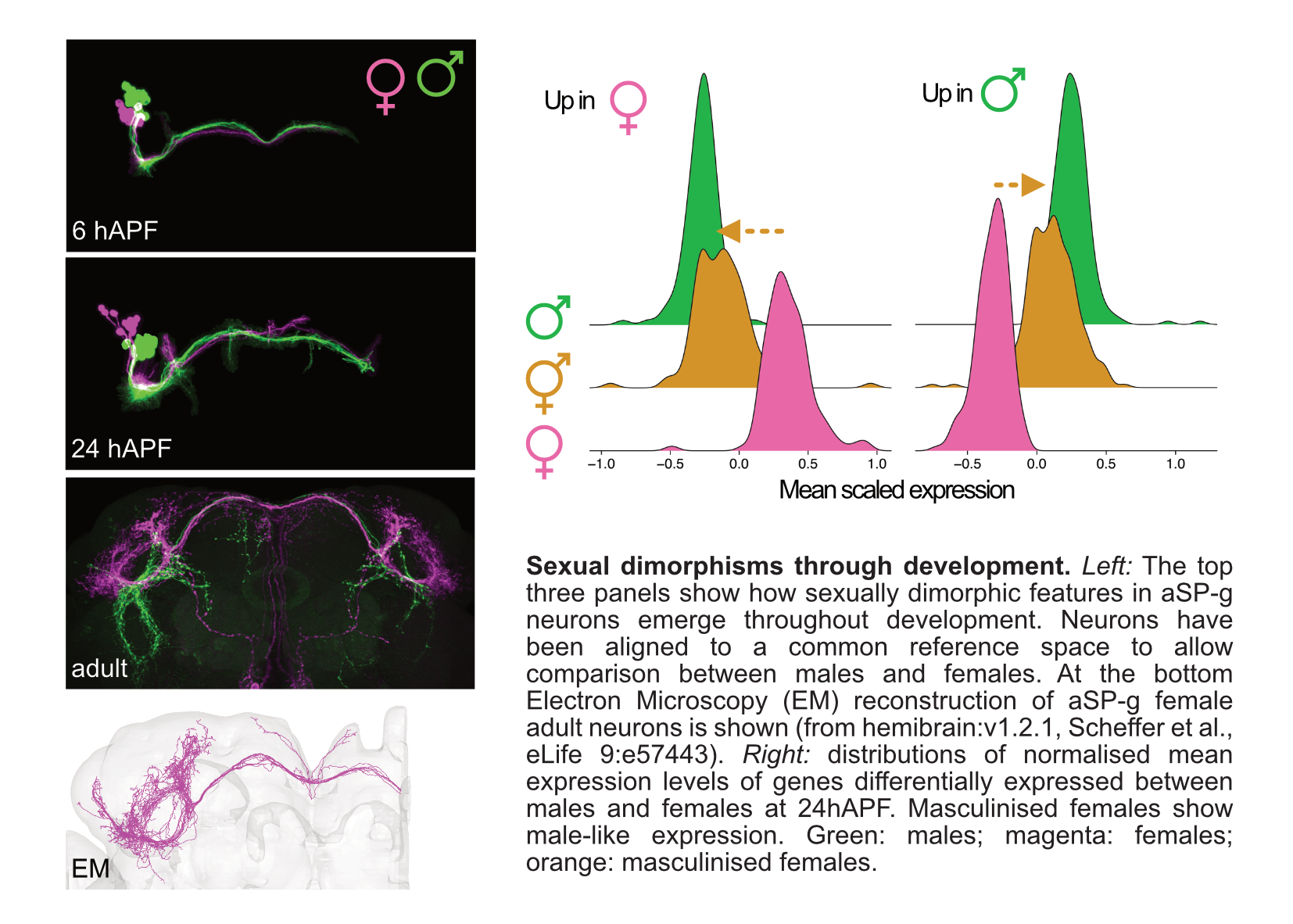
We put particular attention to the study of the development of sexual dimorphisms in the brain, a prime example of how differences in the genome can generate different circuit architectures that lead to different behaviours. As biological sex plays a crucial role in the pathophysiology of many neurodevelopmental disorders, a better understanding of the molecular and developmental basis of physiological male-female differences could help elucidate sex biases in disease conditions.
I am committed to the standards of SAFE Labs, an initiative dedicated to fostering more aware, fair, and equitable research environments.
The lab is actively working to implement all the commitments outlined in the handbook.

Publications
Cachero, Sebastian, Myrto Mitletton, Isabella R. Beckett, Elizabeth C. Marin, Laia Serratosa Capdevila, Marina Gkantia, Haluk Lacin, Gregory S. X. E. Jefferis, and Erika Donà. 2025. “A Developmental Atlas of the Drosophila Nerve Cord Uncovers a Global Temporal Code for Neuronal Identity.” bioRxiv. https://doi.org/10.1101/2025.07.16.664682.
Donà, Erika, James M. Gahan, Petrina Lau, Jana Jeschke, Torben Ott, Katja Reinhard, Chiara Sinigaglia, et al. 2025. “The SAFE Labs Handbook: Community-Driven Commitments for Group Leaders to Improve Lab Culture.” bioRxiv. https://doi.org/10.1101/2025.05.27.655799.
Durdu, Sevi, Murat Iskar, Alicia Roig-Merino, Elisa Gallo, Ezgi Karaca, Andreas Kunze, Erika Doná, Peer Bork, Dirk Schübeler, and Darren Gilmour. 2025. “Nuclear Rerouting of Paracrine Fgf3 in Source Cells Represses Target Genes to Pattern Morphogen Responses.” bioRxiv. https://doi.org/10.1101/2025.07.24.666518.
Taisz, István, Erika Donà, Daniel Münch, Shanice N. Bailey, Billy J. Morris, Kimberly I. Meechan, Katie M. Stevens, et al. 2023. “Generating Parallel Representations of Position and Identity in the Olfactory System.” Cell 186 (12): 2556–73.e22.
Hongjie, L., Janssens J. et al. 2022, Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science 375(6584) (as part of the FCA consortium). DOI: 10.1126/science.abk2432
Donà, E. and Jefferis, G.S.X.E. 2021, Neurodevelopment: Comparative connectomics and the study of circuit assembly. Current Biology 31(9), R452–R454. DOI: 10.1016/j.cub.2021.03.053
Barry, J.D., Donà, E., Gilmour, D., Huber, W. 2016, TimerQuant: a modelling approach to tandem fluorescent timer design and data interpretation for measuring protein turnover in embryos. Development 143, 174-179. DOI: 10.1242/dev.125971
Revenu, C., Streichan, S., Donà, E., Lecaudey, V., Hufnagel, L. and Gilmour, D. 2014, Quantitative cell polarity imaging defines leader-to-follower transitions during collective migration and the key role of microtubule-dependent adherens junction formation. Development 141, 1282–1291. DOI: 10.1242/dev.101675
Donà, E. et al.2013, Directional tissue migration through a self-generated chemokine gradient. Nature 503, 285–289. DOI: 10.1038/nature12635